Efficient first-principles exploration on the physical and chemical space of peptides and saccharides enabled by neural network potentials
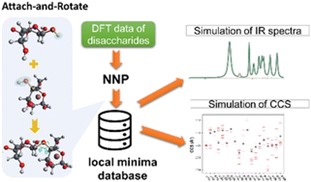
Sampling of the conformational space of peptides and saccharides with first-principles accuracy is critical as such a database provide a solid base to interpret experimental measurements such as Infrared photo-dissociation (IRPD) spectroscopy, ion mobility spectrometry (IMS), and/or collision-induced dissociation (CID). The conformational space of both peptides and saccharides are highly flexible, in which the distinct conformers of mono- and di-saccharide is estimated to be in the order of 103 and 106, respectively1-3. To efficiently explore the diverse conformational space of saccharide without losing accuracy, we developed a multi-level sampling scheme integrating semi-empirical models, density function theory (DFT) and neural network potential (NNP). Preliminary studies on small-size peptides with different protonated sites have also been demonstrated4. Solvation of these molecules can also be simulated with a computational approach that integrate fragment-based methods with NNP5.
References
1) H. T. Phan, P-K Tsou, P-J Hsu, J-L Kuo, Phys. Chem. Chem. Phys. 25, 5817 (2023)
2) P-K Tsou, Hai T. Huynh, H. T. Phan, J-L Kuo, Phys. Chem. Chem. Phys. 25, 3332 (2023)
3) H-T Phan, P-K Tsou, P-J Hsu and J-L Kuo, Phys. Chem. Chem. Phys. 26, 9556 (2024)
4) H-C Dong, P-J Hsu and J-L Kuo, Phys. Chem. Chem. Phys. 26, 11125 (2024)
5) S. Jindal, P-J Hsu, H. T. Phan, P-K Tsou, J-L Kuo, Phys. Chem. Chem. Phys. 24, 27263 (2022)
